Comparison
Plot
Please be aware that only case "thf_base_2018080700" contains
the OUTPUT data directory. This is due
to the size of each case (64G) and data storage constraints.
However validation analysis can be undertaken using the
"thf_base_2018080700"
case.
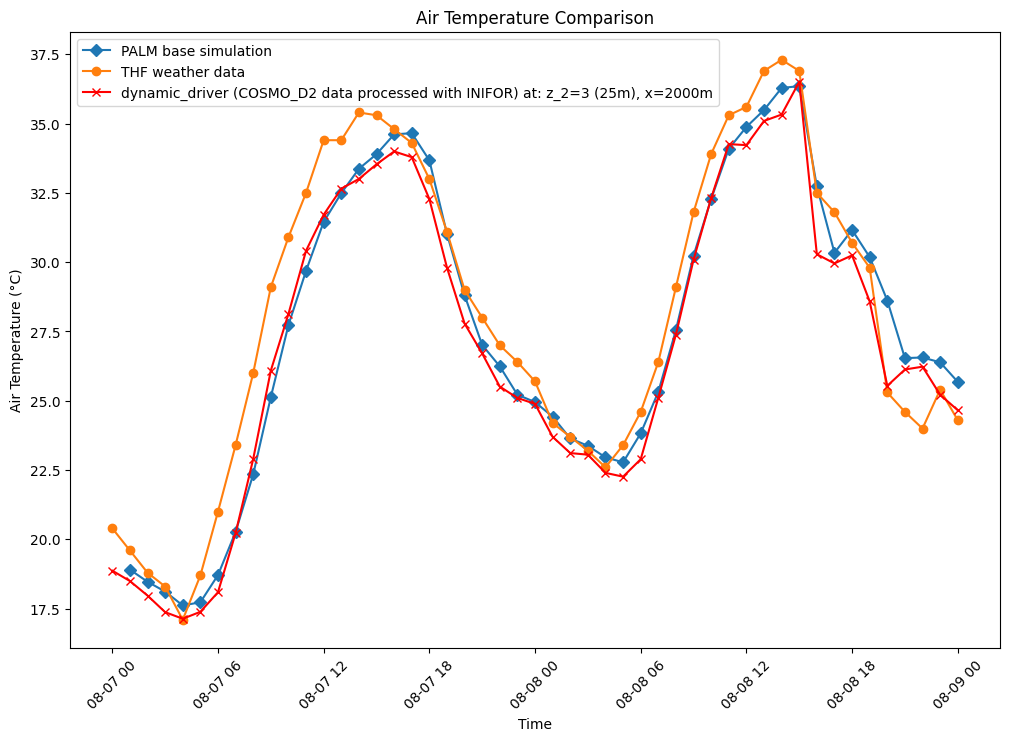
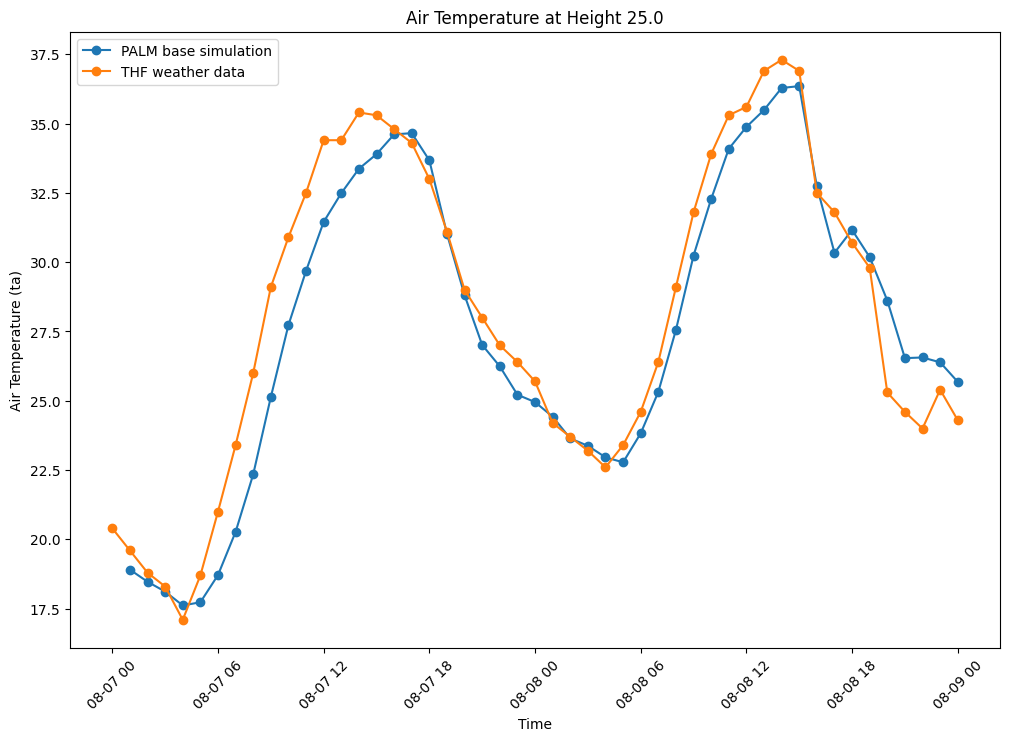
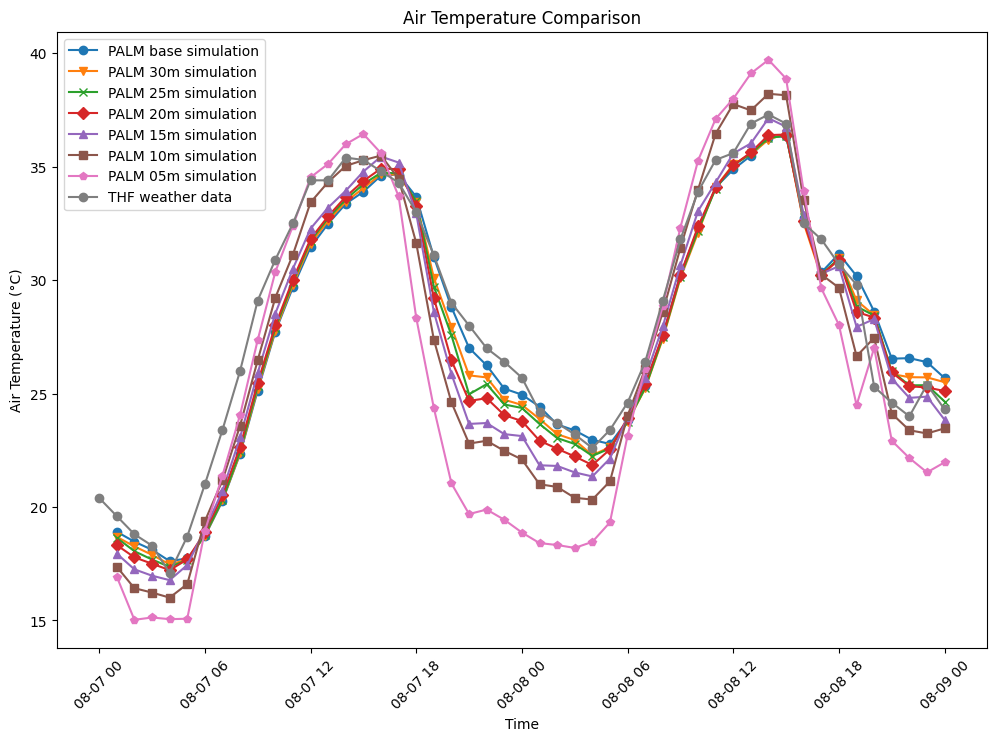
The following figure shows air temperature (degrees celsius) over
two duranial cycles from the date 2018-08-07
00:00:00+00:00 to 2018-08-09 00:00:00+00:00. This time
all tree spacing cases have been included in the plot to cross
compare the results.
Here is a link to the Python code to reproduce the
plot: thf_forest_ta_case_comparison.ipynb
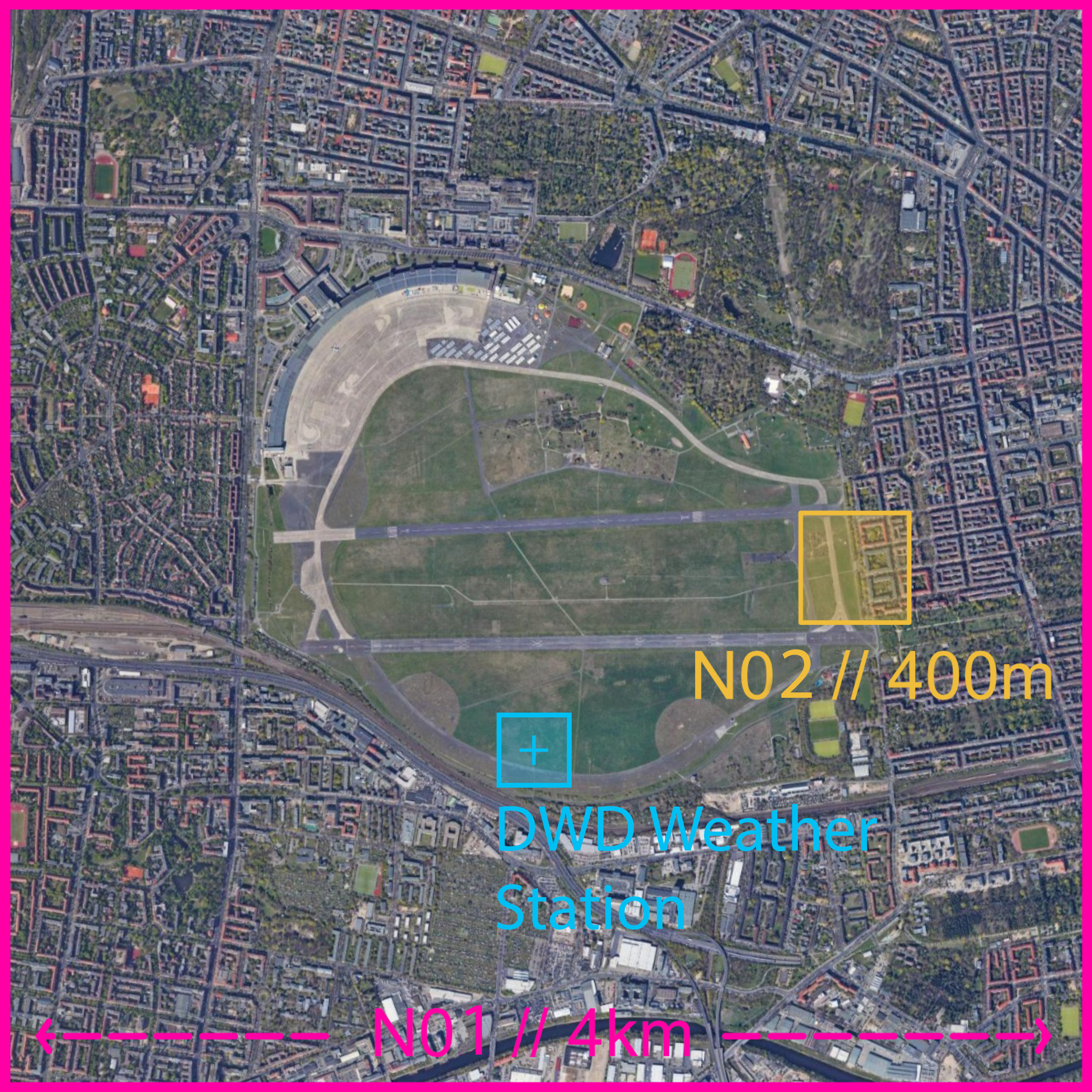
The location of the validation point was set to (DWD weather
station):
x_index = 191 # Grid Cells
y_index = 138 # Grid
Cells
z_index = 3 # Grid
Cells
Discussion
These results must be taken with much caution as full
validation of the PALM Plant Canopy Model (PCM) (specifically 3D
single trees) has not yet been validated. However the validation
of a 2D forest patch with the PALM PCM has
been undertaken - for example Paleri et al. (2023). The following
discussion is just speculation based on preliminary results. I
encourage the PALM developers involved in the PCM to engage in
this discussion to help validate the result.
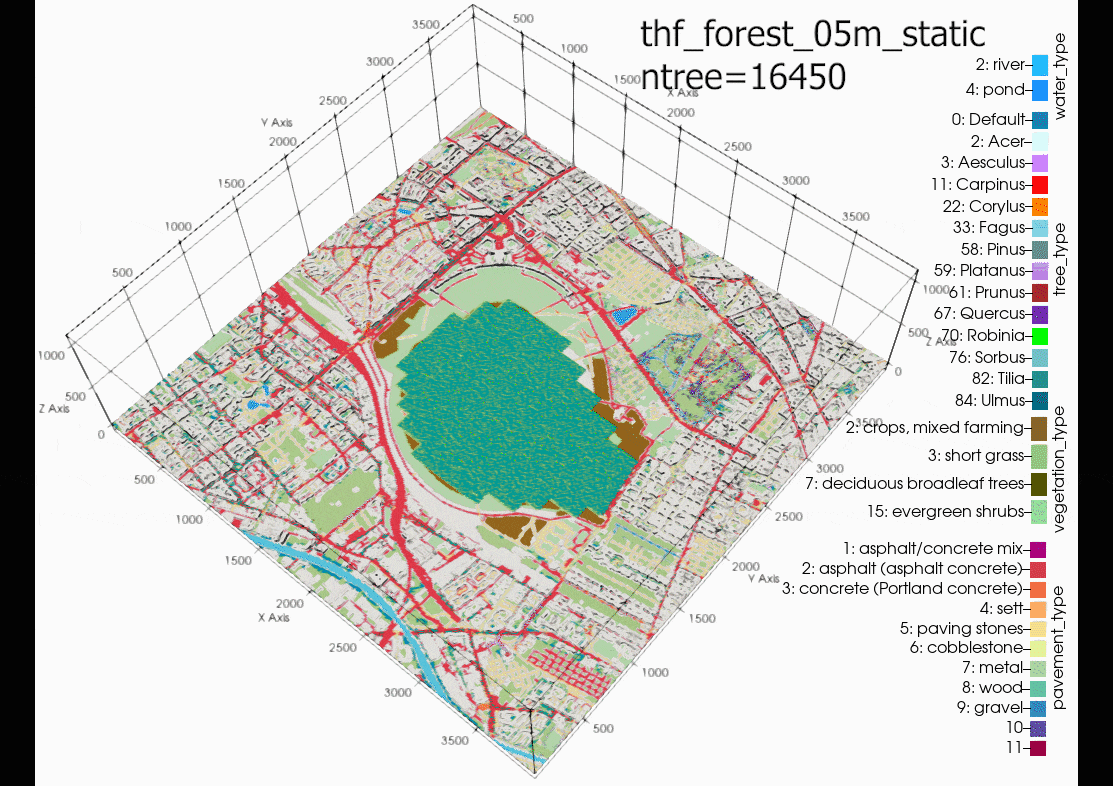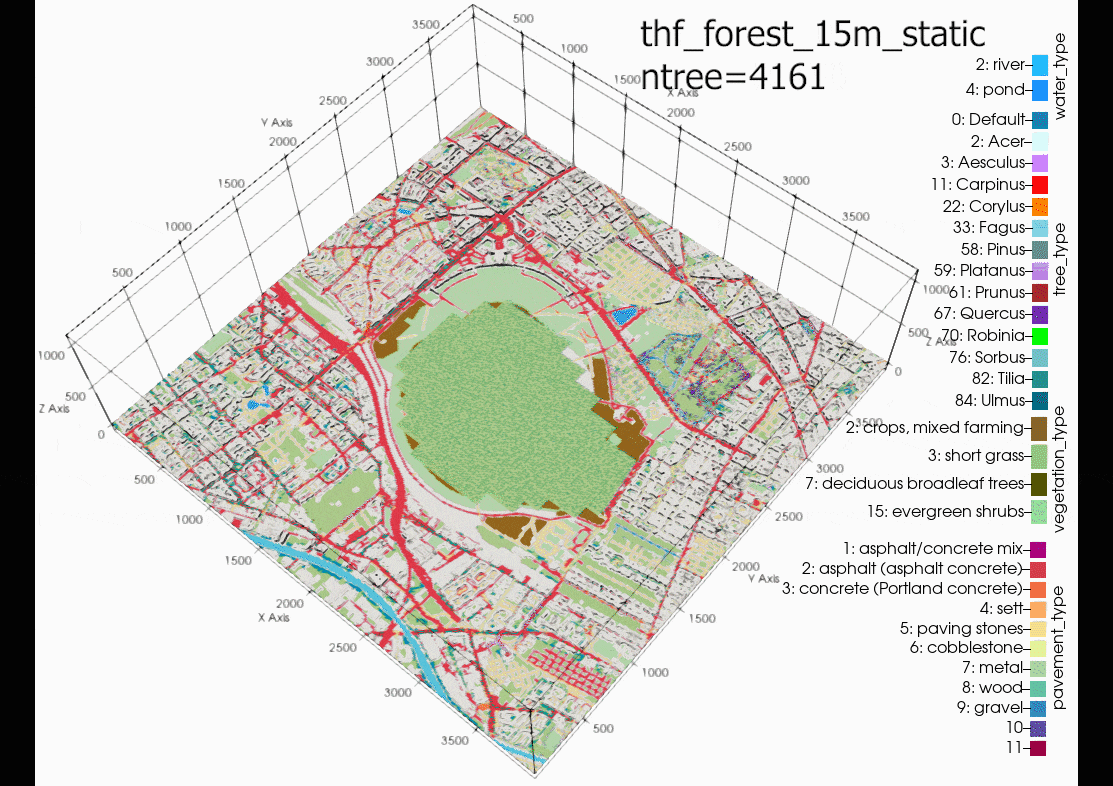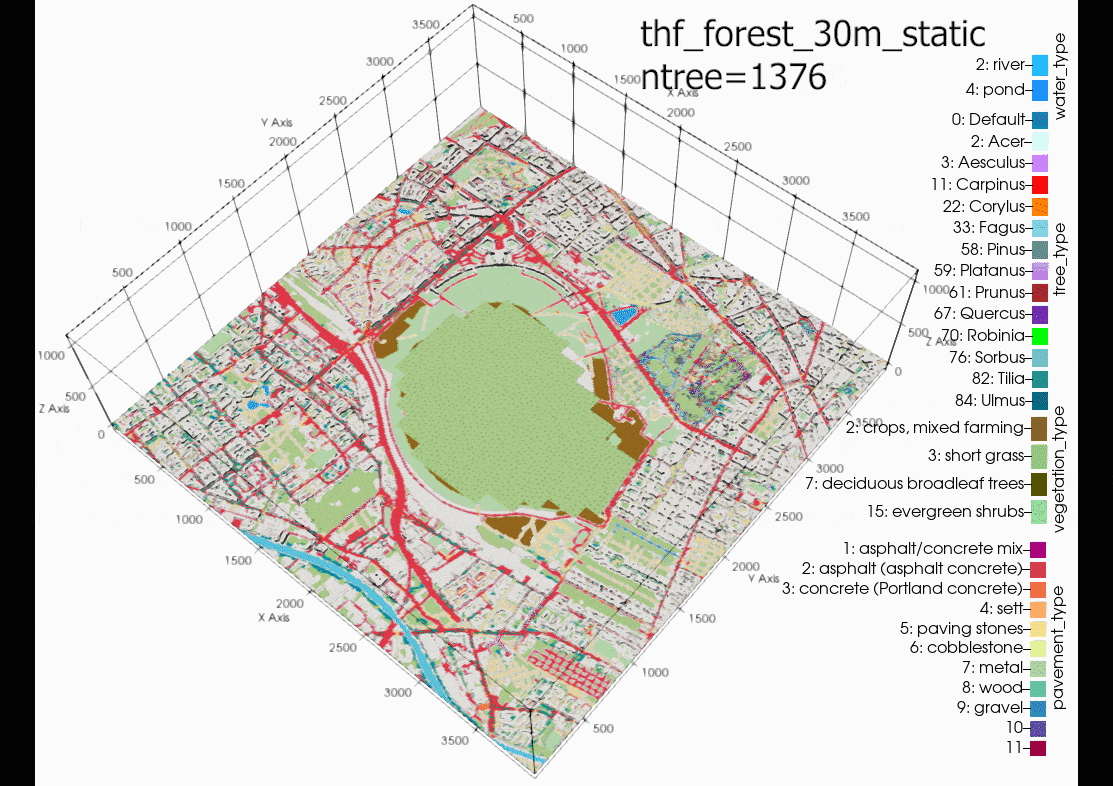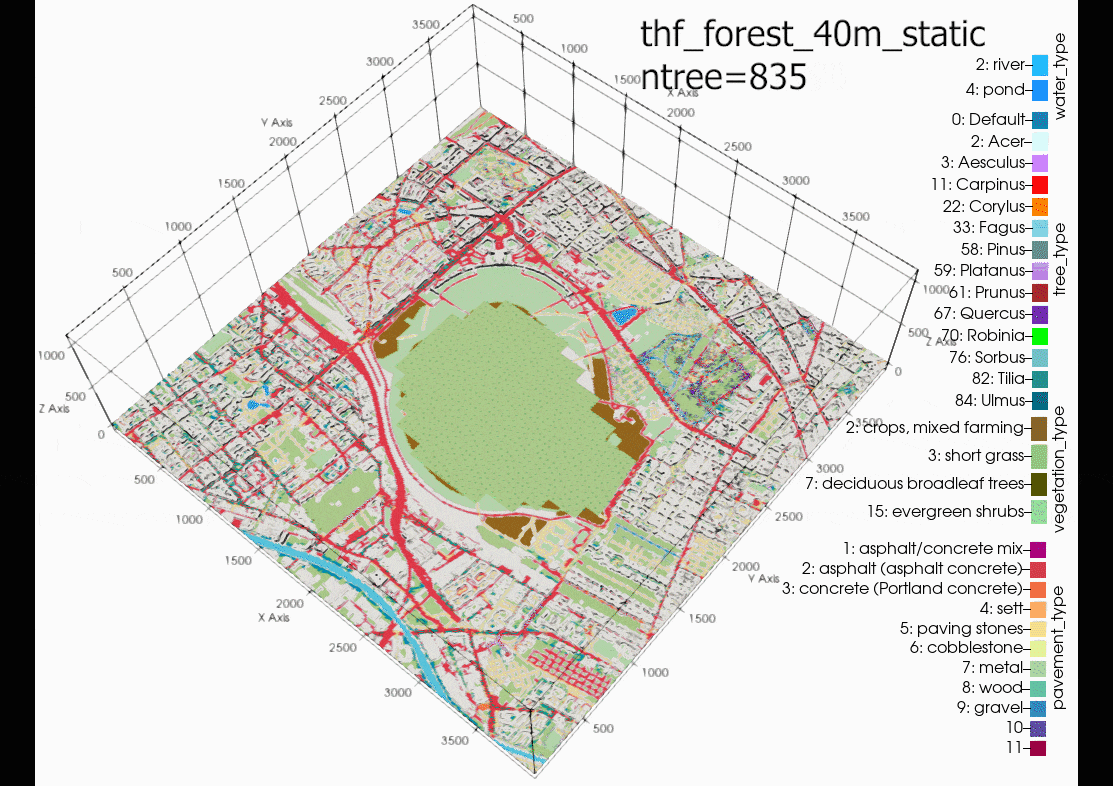
The results show that increasing tree density lowers nighttime
temperature and increases daytime temperatures, compared with the
baseline (blue line) and weather station data (grey line). Some
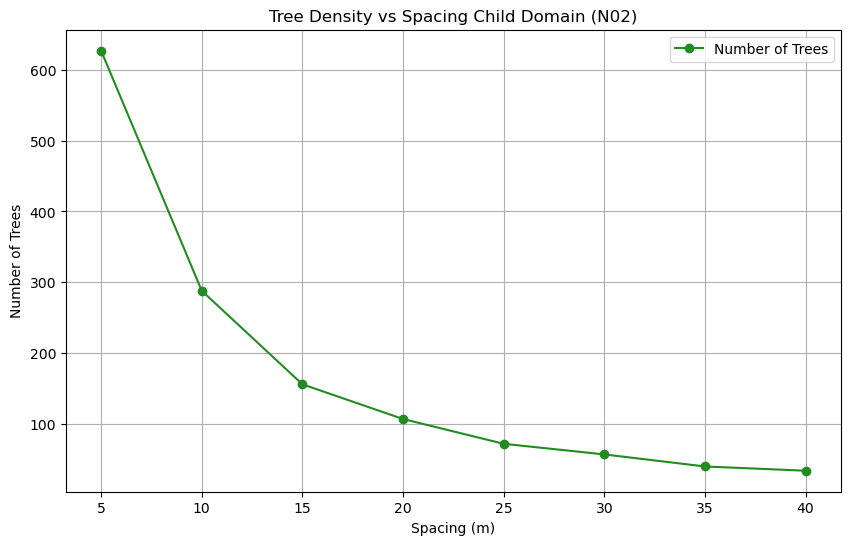
explanation for the exponential nighttime cooling effect, for
example the low temperatures at 08-08-00, case
05m (10m tree spacing trunk to trunk - pink line), in comparison
to the other cases might be due to the exponential increase in
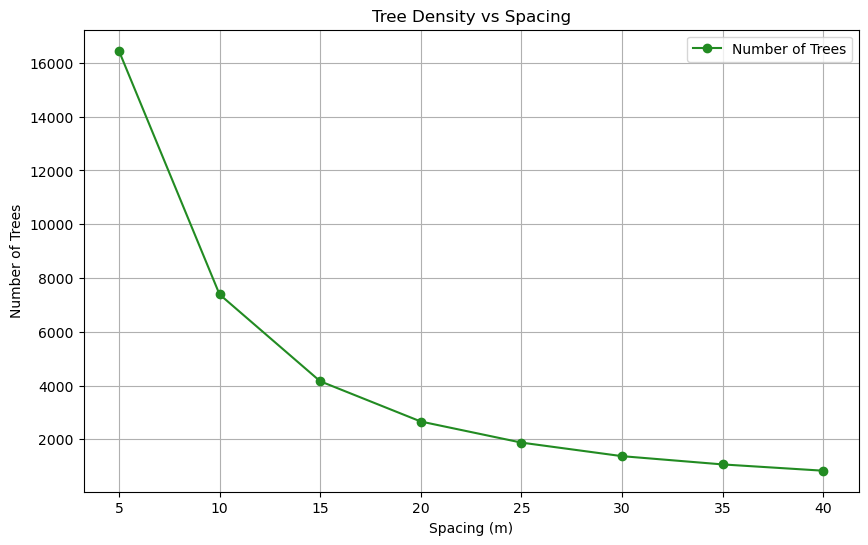
trees, thus more shading and evapotranspiration effect as can be
seen by the exponential curve of trees added at each density in
following figure:
Potential Sources of Error
One potential issue that might cause hightended cooling of
nighttime air temperature in the tree canopy is the following
setting in the p3d file:
&bulk_cloud_parameters
cloud_scheme = 'kessler',
cloud_water_sedimentation = .T.,
Another case without the cloud model is needed to be run in order
to rule out this possible cause of unrealistically low air
temperatures.
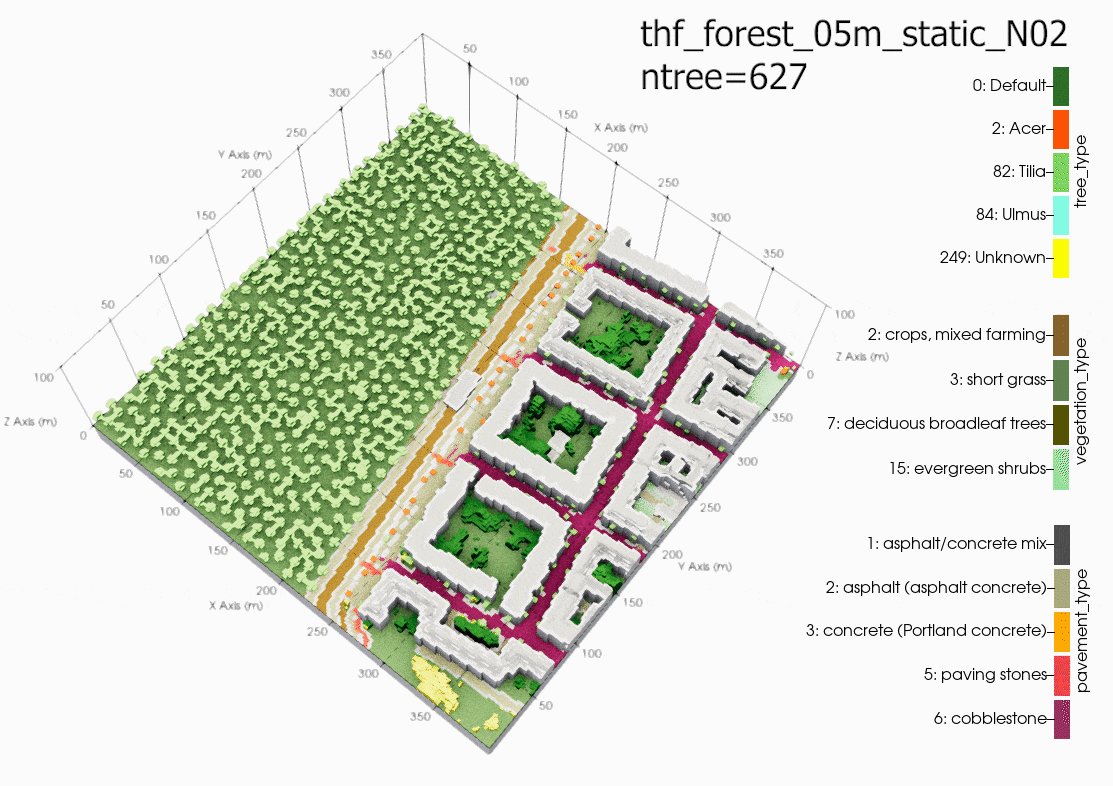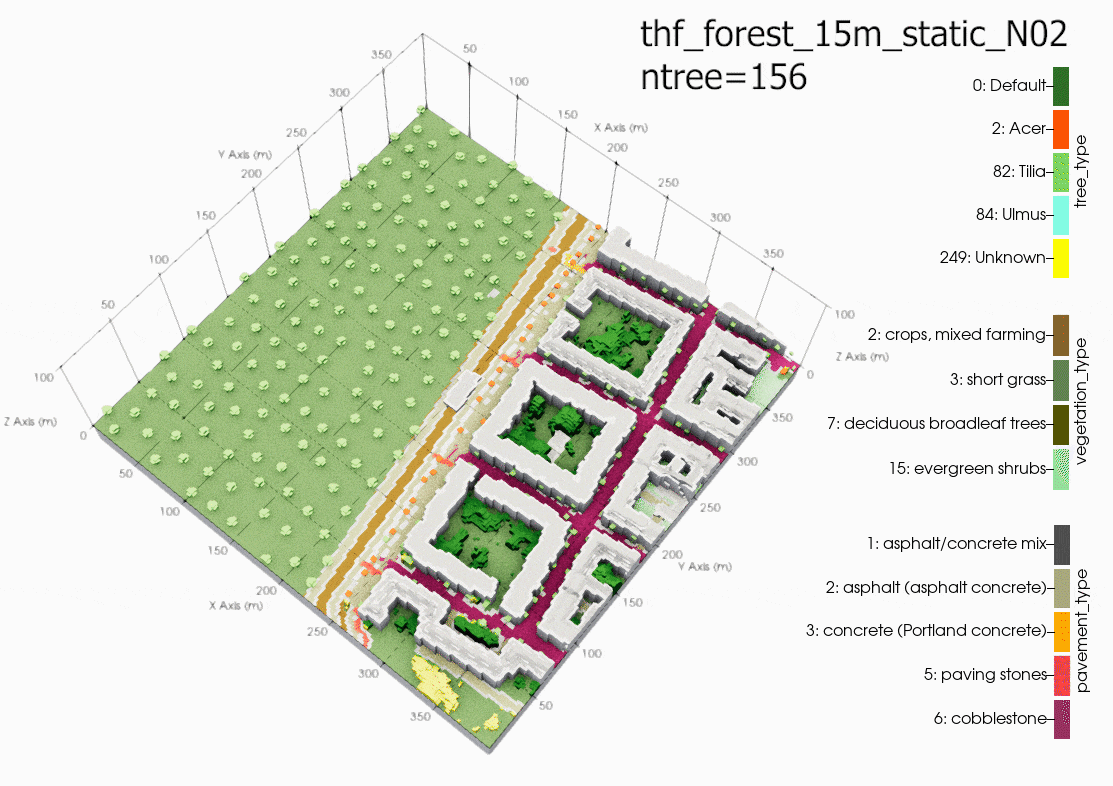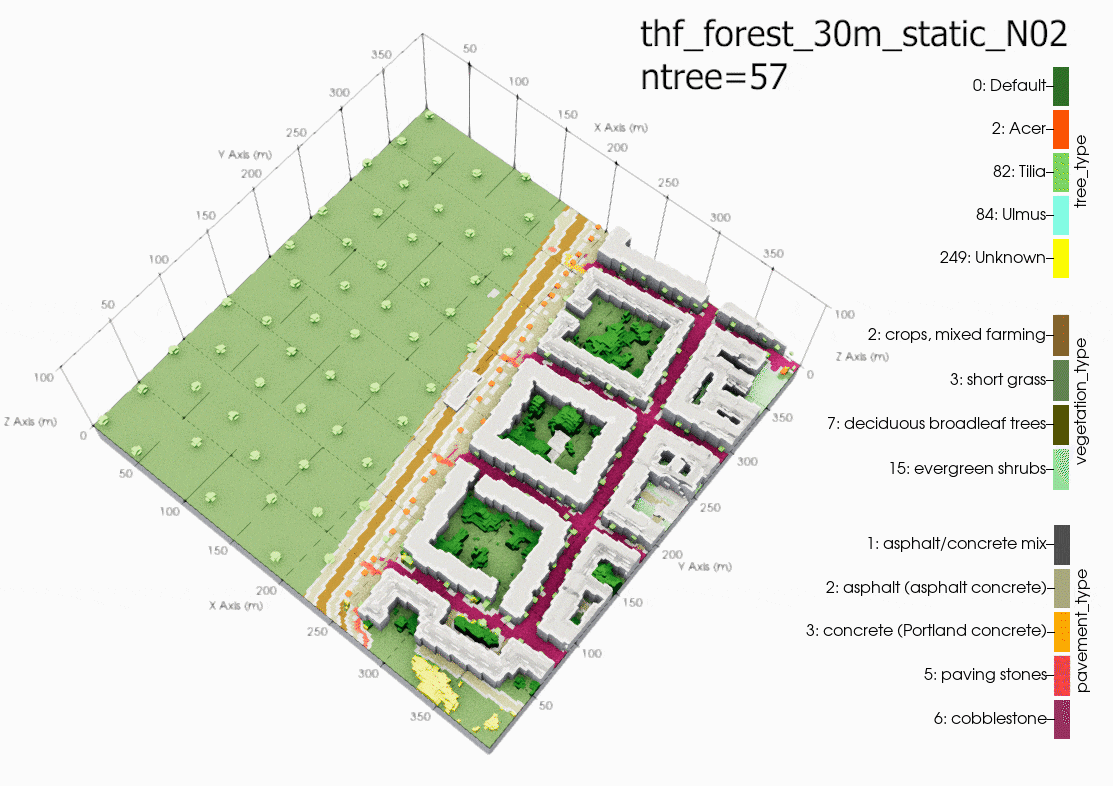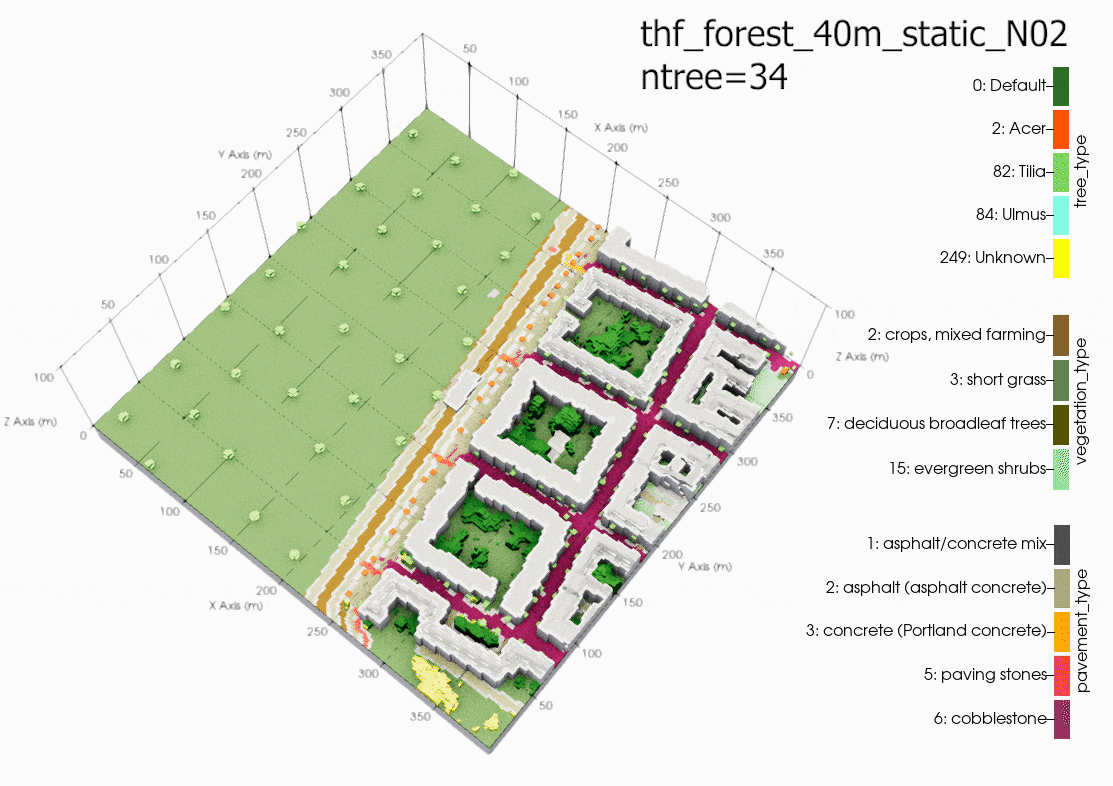
Another source of error could be due to using the single trees in a
domain with large grid spacing (10m in this case). It is
recommended in the PALM documentation to only use single tree with
LAD and BAD profiles when the level of detail (LOD1) is less than a
1m grid resolution. Although, the same air temperatures can be
found in the child domain (N02). Suggesting that grid sensitivity
to the single trees in PALM is not the main cause of error. This
cannot be proven until another case is run with a 1m grid
resolution in the child domain.
Specifying the full range of tree options via the
palm_csd Python script is needed to make sure the
trees in the study are specified correctly. Below is a list of
possible single tree options available:
|
Index
|
|=Species
|
Shape
|
Crown height/width ratio(*)
|
Crown diameter (m)
|
Height (m)
|
LAI summer(*)
|
LAI winter(*)
|
Height of maximum LAD (m)(*)
|
LAD/BAD ratio(*)
|
DBH (m)
|
|
0
|
Default
|
1.0
|
1.0
|
4.0
|
12.0
|
3.0
|
0.8
|
0.6
|
0.025
|
0.35
|
|
1
|
Abies
|
3.0
|
1.0
|
4.0
|
12.0
|
3.0
|
0.8
|
0.6
|
0.025
|
0.80
|
|
2
|
Acer
|
1.0
|
1.0
|
7.0
|
12.0
|
3.0
|
0.8
|
0.6
|
0.025
|
0.80
|
Currently only the following tree options have been switched on as
you can see in this example CSV array:
i,j,height,type,tree_dia,trunk_dia,tree_lai
123,229,20,82,10,1,3
Crown height/width
ratio(*), DBH, Height of maximum
LAD (m)(*), LAD/BAD ratio(*)
and DBH (m) are still yet to be
utilised!
Feedback from the PALM developers and personnel
responsible for maintaining the PALM Plant Canopy Model and
palm_csd Python Processor would be most welcome. I believe that a
thorough investigation and validation of the PMC would also be
beneficial to the PALM community and scientific
users.